Introduction to Medusa¶
Loading an example ensemble and inspecting its parts¶
In medusa, ensembles of genome-scale metabolic network
reconstructions (GENREs) are represented using the medusa.Ensemble
class. To demonstrate the functionality and attributes of this class,
we’ll, load a test ensemble. Here, we use a function that takes the E.
coli core metabolism reconstruction from cobrapy and randomly removes
components to generate ensemble members.
In [1]:
import medusa
from medusa.test.test_ensemble import construct_textbook_ensemble
example_ensemble = construct_textbook_ensemble()
Each Ensemble has three key attributes that specify the structure of
the ensemble, which we’ll describe below. This schematic also summarizes
the structure of Ensemble and how each attribute relates to cobrapy
objects:
In [2]:
from IPython.display import Image
Image(filename='medusa_structure.png', width=500)
Out[2]:
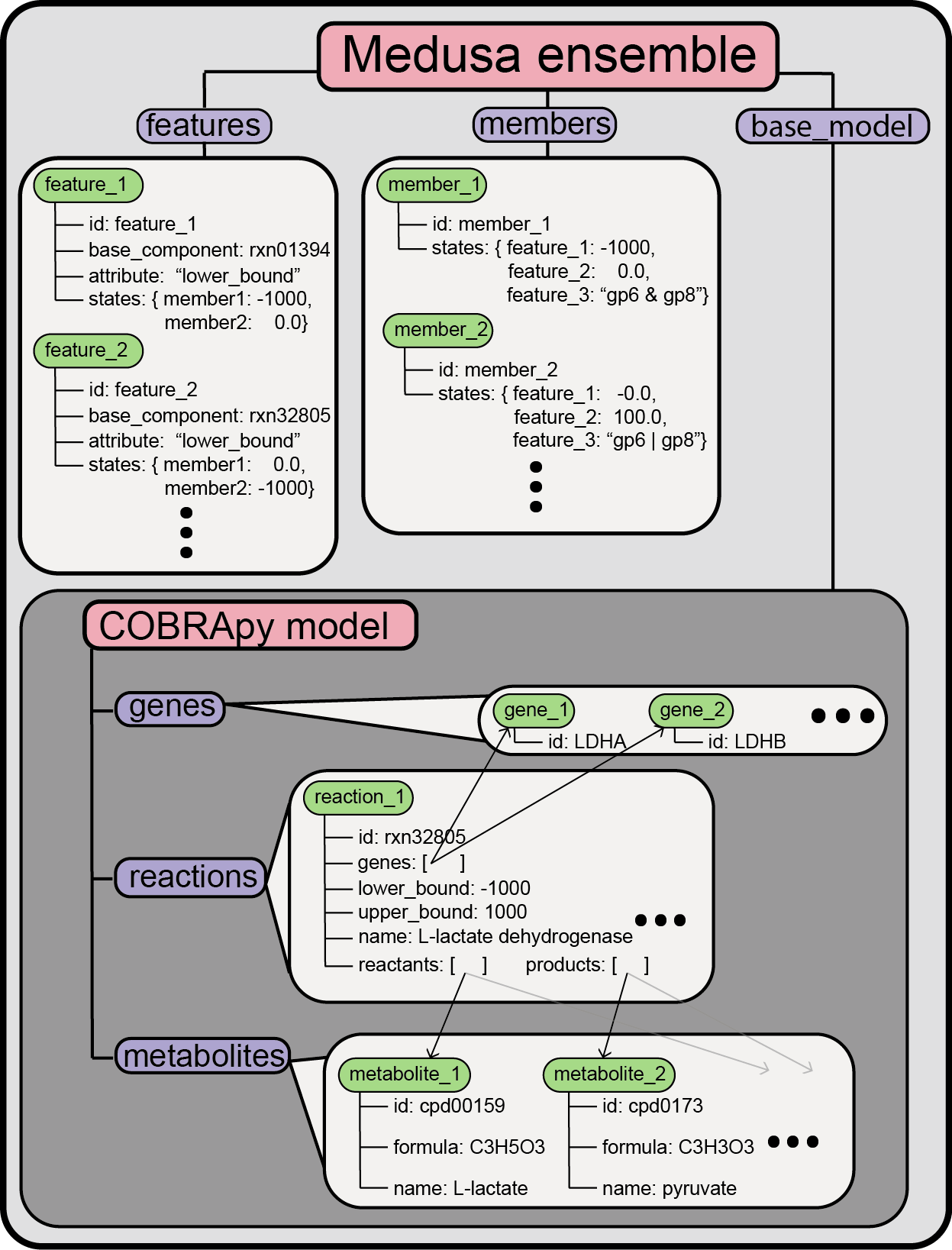
Components of an ensemble: base_model¶
The first is the base_model, which is a cobra.Model object that
represents all the possible states of an individual member within the
ensemble. Any reaction, metabolite, or gene that is only present in a
subset of ensemble members will be present in the base_model for an
Ensemble. You can inspect the base_model and manipulate it just
like any other cobra.Model object.
In [3]:
extracted_base_model = example_ensemble.base_model
extracted_base_model
Out[3]:
| Name | first_textbook |
| Memory address | 0x07efcac754f98 |
| Number of metabolites | 72 |
| Number of reactions | 95 |
| Objective expression | 1.0*Biomass_Ecoli_core - 1.0*Biomass_Ecoli_core_reverse_2cdba |
| Compartments | cytosol, extracellular |
Components of an ensemble: members¶
The second attribute that each Ensemble has is a structure called
members. Ensemble.members maps an identifier for each individual
GENRE in the ensemble to a medusa.Member object, which holds
information about a single member (where a “single member” is an
individual GENRE within an ensemble).
Ensemble.members is represented by a custom class implemented in
cobrapy called a
DictList,
which is essentially a standard dictionary in python that can also be
accessed using integer indices like a list (e.g. dictlist[0] returns the
first element in the dictlist).
In [4]:
# looks like a list when we print it
example_ensemble.members
Out[4]:
[<Member first_textbook at 0x7efcac983be0>,
<Member second_textbook at 0x7efcac983ef0>]
In [5]:
# Get the first member with integer indexing
first_member = example_ensemble.members[0]
Each Member within the Ensemble.members DictList has a handful
of attributes as well. You can check the ensemble that the member
belongs to, the id of the member, and the network states for that member
(we’ll discuss states more below).
In [6]:
print(first_member.ensemble)
print(first_member.id)
print(first_member.states)
textbook_ensemble
first_textbook
{<Feature ACONTb_lower_bound at 0x7efcac761e10>: -1000.0, <Feature ACKr_lower_bound at 0x7efcac91d128>: 0.0, <Feature ACt2r_upper_bound at 0x7efcac91db38>: 1000.0, <Feature ACt2r_lower_bound at 0x7efcac91dd68>: -1000.0, <Feature ACKr_upper_bound at 0x7efcac91d668>: 0.0, <Feature ACALDt_upper_bound at 0x7efcac91d198>: 0.0, <Feature ACALDt_lower_bound at 0x7efcac91d390>: 0.0, <Feature ACONTb_upper_bound at 0x7efcac91def0>: 1000.0}
Components of an ensemble: features¶
The states printed above are directly connected to the third attribute
that Ensemble contains, Ensemble.features, which is also a
DictList object. Ensemble.features contains medusa.Feature
entries, which specify the components of the Ensemble.base_model
that vary across the entire ensemble.
In [7]:
example_ensemble.features
Out[7]:
[<Feature ACALDt_lower_bound at 0x7efcac91d390>,
<Feature ACALDt_upper_bound at 0x7efcac91d198>,
<Feature ACKr_lower_bound at 0x7efcac91d128>,
<Feature ACKr_upper_bound at 0x7efcac91d668>,
<Feature ACONTb_lower_bound at 0x7efcac761e10>,
<Feature ACONTb_upper_bound at 0x7efcac91def0>,
<Feature ACt2r_lower_bound at 0x7efcac91dd68>,
<Feature ACt2r_upper_bound at 0x7efcac91db38>]
Here, we see that this Ensemble has 8 features. Each Feature
object specifies a network component that has a variable parameter value
in at least one member of the ensemble (e.g. at least one ensemble
member is missing the reaction).
In this case, there are features for 4 reactions,
ACALDt,ACKr,ACONTb, and ACt2r. There are two
Feature objects for each reaction, corresponding to the lower and
upper bound for that reaction. A feature will be generated for any
component of a cobra.Model (e.g. Reaction, Gene) that has an
attribute value (e.g. Reaction.lower_bound,
Reaction.gene_reaction_rule) that varies across the ensemble. As you
can see from this result, a feature is created at the level of the
specific attribute that varies, not the model component (e.g. we
created a Feature for each bound of each Reaction, not for the
Reaction objects themselves).
This information can be inferred from feature ID
(medusa.Feature.id), but each Feature also has a set of
attributes that encode the information. Some useful attributes,
described in the order printed below: getting the Ensemble that the
Feature belongs to, the component in the Ensemble.base_model
that the Feature describes, the attribute of the component in the
Ensemble.base_model whose value the Feature specifies, and the
ID of the Feature:
In [8]:
first_feature = example_ensemble.features[0]
print(first_feature.ensemble)
print(first_feature.base_component)
print(first_feature.component_attribute)
print(first_feature.id)
textbook_ensemble
ACALDt: acald_e <=> acald_c
lower_bound
ACALDt_lower_bound
Just as each member has an attribute, states, that returns the
value of every feature for that member, each feature has a
states dictionary that maps each member.id to the value of the
feature in the corresponding member, e.g.:
In [9]:
print(first_feature.states)
{'second_textbook': -1000.0, 'first_textbook': 0.0}
Strategies for getting information about an ensemble and its members¶
Where possible, we use conventions from cobrapy for accessing
information about attributes. In cobrapy, the Model object has
multiple containers in the form of DictLists:
Model.reactions,Model.metabolites,Model.genes.
Equivalently in medusa, each Ensemble has similarly constructed
containers: Ensemble.members and Ensemble.features.
As such, information about specific Member and Feature objects
can be accessed just like Reaction, Metabolite, and Gene
objects in cobrapy:
In [10]:
# Remember, our Ensemble holds a normal cobrapy Model in base_model
extracted_base_model = example_ensemble.base_model
# Accessing object by id is common in cobrapy
rxn = extracted_base_model.reactions.get_by_id('ACALDt')
# We can do the same thing for features:
feat = example_ensemble.features.get_by_id('ACALDt_lower_bound')
print(rxn)
print(feat.base_component)
print(feat.component_attribute)
# And for members:
memb = example_ensemble.members.get_by_id('first_textbook')
print('\nHere are the states for this member:')
print(memb.states)
ACALDt: acald_e <=> acald_c
ACALDt: acald_e <=> acald_c
lower_bound
Here are the states for this member:
{<Feature ACONTb_lower_bound at 0x7efcac761e10>: -1000.0, <Feature ACKr_lower_bound at 0x7efcac91d128>: 0.0, <Feature ACt2r_upper_bound at 0x7efcac91db38>: 1000.0, <Feature ACt2r_lower_bound at 0x7efcac91dd68>: -1000.0, <Feature ACKr_upper_bound at 0x7efcac91d668>: 0.0, <Feature ACALDt_upper_bound at 0x7efcac91d198>: 0.0, <Feature ACALDt_lower_bound at 0x7efcac91d390>: 0.0, <Feature ACONTb_upper_bound at 0x7efcac91def0>: 1000.0}
These DictList objects are all
iterables,
meaning that any python operation that acts on an iterable can take them
as input. This is often convenient when working with either cobrapy
Models or medusa Ensembles. For example, suppose we are
interested in getting the list of all components described by features
in the Ensemble:
In [11]:
components = []
for feat in example_ensemble.features:
components.append(feat.base_component)
print(components)
# or, use the one-liner which gives the same result:
components = [feat.base_component for feat in example_ensemble.features]
print(components)
[<Reaction ACALDt at 0x7efcac66f0b8>, <Reaction ACALDt at 0x7efcac66f0b8>, <Reaction ACKr at 0x7efcac66f128>, <Reaction ACKr at 0x7efcac66f128>, <Reaction ACONTb at 0x7efcac758b00>, <Reaction ACONTb at 0x7efcac758b00>, <Reaction ACt2r at 0x7efcac7587f0>, <Reaction ACt2r at 0x7efcac7587f0>]
[<Reaction ACALDt at 0x7efcac66f0b8>, <Reaction ACALDt at 0x7efcac66f0b8>, <Reaction ACKr at 0x7efcac66f128>, <Reaction ACKr at 0x7efcac66f128>, <Reaction ACONTb at 0x7efcac758b00>, <Reaction ACONTb at 0x7efcac758b00>, <Reaction ACt2r at 0x7efcac7587f0>, <Reaction ACt2r at 0x7efcac7587f0>]