Performing ensemble simulations¶
With a functional Ensemble in hand, you’re ready to perform
simulations. In medusa, most simulations are performed by setting
the model structure to represent an individual member, using cobrapy
functions for the actual simulation, then repeating for all or many
ensemble members.
Ensemble Flux Balance Analysis¶
Flux balance analysis (FBA) is one of the most widely used techniques in systems biology. See What is flux balance analysis? for an introduction to FBA, and the cobrapy documentation to see how FBA is performed with a single model.
When using medusa for FBA, the environmental conditions and
objective function should be specified in ensemble.base_model, just
as if it were a normal cobrapy Model:
In [1]:
import medusa
from medusa.test import create_test_ensemble
ensemble = create_test_ensemble("Staphylococcus aureus")
In [2]:
ensemble.base_model.objective.expression
Out[2]:
1.0*bio1 - 1.0*bio1_reverse_b18f7
The current objective function is the biomass reaction (bio1)–to
change this, just set the objective to another reaction. Let’s change
the objective to CO2 exchange, then change it back to biomass
production:
In [3]:
ensemble.base_model.objective = 'EX_cpd00011_e'
print(ensemble.base_model.objective.expression)
ensemble.base_model.objective = 'bio1'
print(ensemble.base_model.objective.expression)
1.0*EX_cpd00011_e - 1.0*EX_cpd00011_e_reverse_896eb
1.0*bio1 - 1.0*bio1_reverse_b18f7
Similarly, you can manipulate the environmental conditions as in
cobrapy. The base model for this example ensemble is from ModelSEED, so
exchange reactions are specified with the 'EX_' prefix, followed by
the metabolite id. Let’s take a look at the exchange reactions that are
currently open:
In [4]:
medium = ensemble.base_model.medium
medium
Out[4]:
{'EX_cpd00001_e': 1000.0,
'EX_cpd00007_e': 1000.0,
'EX_cpd00009_e': 1000.0,
'EX_cpd00010_e': 1000.0,
'EX_cpd00011_e': 1000.0,
'EX_cpd00012_e': 1000.0,
'EX_cpd00013_e': 1000,
'EX_cpd00023_e': 1000.0,
'EX_cpd00024_e': 1000.0,
'EX_cpd00027_e': 1000.0,
'EX_cpd00028_e': 1000.0,
'EX_cpd00029_e': 1000,
'EX_cpd00030_e': 1000.0,
'EX_cpd00033_e': 1000.0,
'EX_cpd00034_e': 1000.0,
'EX_cpd00035_e': 1000.0,
'EX_cpd00039_e': 1000.0,
'EX_cpd00041_e': 1000.0,
'EX_cpd00047_e': 1000.0,
'EX_cpd00048_e': 1000.0,
'EX_cpd00051_e': 1000.0,
'EX_cpd00053_e': 1000.0,
'EX_cpd00054_e': 1000.0,
'EX_cpd00058_e': 1000.0,
'EX_cpd00060_e': 1000.0,
'EX_cpd00063_e': 1000.0,
'EX_cpd00064_e': 1000.0,
'EX_cpd00066_e': 1000.0,
'EX_cpd00067_e': 1000.0,
'EX_cpd00069_e': 1000.0,
'EX_cpd00072_e': 1000,
'EX_cpd00073_e': 1000.0,
'EX_cpd00075_e': 1000.0,
'EX_cpd00076_e': 1000.0,
'EX_cpd00079_e': 1000,
'EX_cpd00080_e': 1000.0,
'EX_cpd00082_e': 1000.0,
'EX_cpd00092_e': 1000.0,
'EX_cpd00094_e': 1000,
'EX_cpd00098_e': 1000.0,
'EX_cpd00099_e': 1000.0,
'EX_cpd00100_e': 1000.0,
'EX_cpd00104_e': 1000.0,
'EX_cpd00105_e': 1000.0,
'EX_cpd00117_e': 1000.0,
'EX_cpd00119_e': 1000.0,
'EX_cpd00122_e': 1000.0,
'EX_cpd00129_e': 1000.0,
'EX_cpd00130_e': 1000.0,
'EX_cpd00137_e': 1000.0,
'EX_cpd00138_e': 1000.0,
'EX_cpd00141_e': 1000,
'EX_cpd00142_e': 1000,
'EX_cpd00149_e': 1000.0,
'EX_cpd00159_e': 1000.0,
'EX_cpd00179_e': 1000.0,
'EX_cpd00182_e': 1000.0,
'EX_cpd00184_e': 1000.0,
'EX_cpd00205_e': 1000.0,
'EX_cpd00208_e': 1000.0,
'EX_cpd00220_e': 1000.0,
'EX_cpd00222_e': 1000.0,
'EX_cpd00232_e': 1000,
'EX_cpd00244_e': 1000.0,
'EX_cpd00246_e': 1000.0,
'EX_cpd00249_e': 1000.0,
'EX_cpd00254_e': 1000.0,
'EX_cpd00264_e': 1000.0,
'EX_cpd00268_e': 1000.0,
'EX_cpd00276_e': 1000.0,
'EX_cpd00277_e': 1000.0,
'EX_cpd00305_e': 1000.0,
'EX_cpd00309_e': 1000.0,
'EX_cpd00314_e': 1000.0,
'EX_cpd00320_e': 1000,
'EX_cpd00322_e': 1000.0,
'EX_cpd00355_e': 1000.0,
'EX_cpd00367_e': 1000.0,
'EX_cpd00393_e': 1000.0,
'EX_cpd00396_e': 1000,
'EX_cpd00412_e': 1000.0,
'EX_cpd00438_e': 1000.0,
'EX_cpd00492_e': 1000,
'EX_cpd00531_e': 1000.0,
'EX_cpd00540_e': 1000.0,
'EX_cpd00550_e': 1000.0,
'EX_cpd00588_e': 1000.0,
'EX_cpd00637_e': 1000.0,
'EX_cpd00654_e': 1000.0,
'EX_cpd00681_e': 1000.0,
'EX_cpd00709_e': 1000,
'EX_cpd00794_e': 1000.0,
'EX_cpd00971_e': 1000.0,
'EX_cpd01012_e': 1000.0,
'EX_cpd01030_e': 1000.0,
'EX_cpd01080_e': 1000.0,
'EX_cpd01171_e': 1000.0,
'EX_cpd01262_e': 1000.0,
'EX_cpd01293_e': 1000,
'EX_cpd01307_e': 1000,
'EX_cpd01329_e': 1000.0,
'EX_cpd01914_e': 1000.0,
'EX_cpd03279_e': 1000.0,
'EX_cpd03561_e': 1000,
'EX_cpd03696_e': 1000.0,
'EX_cpd03724_e': 1000.0,
'EX_cpd03725_e': 1000.0,
'EX_cpd04097_e': 1000.0,
'EX_cpd05158_e': 1000,
'EX_cpd05264_e': 1000,
'EX_cpd08305_e': 1000.0,
'EX_cpd08306_e': 1000.0,
'EX_cpd10515_e': 1000.0,
'EX_cpd10516_e': 1000.0,
'EX_cpd11576_e': 1000.0,
'EX_cpd11594_e': 1000,
'EX_cpd11597_e': 1000.0,
'EX_cpd15584_e': 1000,
'EX_cpd19001_e': 1000}
That’s a lot of open exchange reactions! Let’s make them a bit more
realistic for an in vitro situation. We’ll load a file specifying the
base composition of the media in biolog single C/N growth conditions,
and set the media conditions to reflect that. The base composition is
missing a carbon source, so we’ll enable uptake of glucose. In the
medium dictionary, the numbers for each exchange reaction are uptake
rates. If you inspect the actual exchange reactions, you will find that
the equivalent to an uptake rate of 1000 units is a lower bound of
-1000, because our exchange reactions are defined with the boundary
metabolite as the reactant, e.g. cpd00182_e -->.
In [5]:
import pandas as pd
biolog_base = pd.read_csv("../medusa/test/data/biolog_base_composition.csv", sep=",")
biolog_base
Out[5]:
| Name | ID | |
|---|---|---|
| 0 | H2O | cpd00001_e |
| 1 | O2 | cpd00007_e |
| 2 | Phosphate | cpd00009_e |
| 3 | CO2 | cpd00011_e |
| 4 | NH3 | cpd00013_e |
| 5 | Mn2+ | cpd00030_e |
| 6 | Zn2+ | cpd00034_e |
| 7 | Sulfate | cpd00048_e |
| 8 | Cu2+ | cpd00058_e |
| 9 | Ca2+ | cpd00063_e |
| 10 | H+ | cpd00067_e |
| 11 | Cl- | cpd00099_e |
| 12 | Co2+ | cpd00149_e |
| 13 | K+ | cpd00205_e |
| 14 | Mg | cpd00254_e |
| 15 | Na+ | cpd00971_e |
| 16 | Fe2+ | cpd10515_e |
| 17 | fe3 | cpd10516_e |
| 18 | Heme | cpd00028_e |
| 19 | H2S2O3 | cpd00268_e |
In [6]:
# convert the biolog base to a dictionary, which we can use to set ensemble.base_model.medium directly.
biolog_base = {'EX_'+component:1000 for component in biolog_base['ID']}
# add glucose uptake to the new medium dictionary
biolog_base['EX_cpd00182_e'] = 10
# Set the medium on the base model
ensemble.base_model.medium = biolog_base
ensemble.base_model.medium
Out[6]:
{'EX_cpd00001_e': 1000,
'EX_cpd00007_e': 1000,
'EX_cpd00009_e': 1000,
'EX_cpd00011_e': 1000,
'EX_cpd00013_e': 1000,
'EX_cpd00028_e': 1000,
'EX_cpd00030_e': 1000,
'EX_cpd00034_e': 1000,
'EX_cpd00048_e': 1000,
'EX_cpd00058_e': 1000,
'EX_cpd00063_e': 1000,
'EX_cpd00067_e': 1000,
'EX_cpd00099_e': 1000,
'EX_cpd00149_e': 1000,
'EX_cpd00182_e': 10,
'EX_cpd00205_e': 1000,
'EX_cpd00254_e': 1000,
'EX_cpd00268_e': 1000,
'EX_cpd00971_e': 1000,
'EX_cpd10515_e': 1000,
'EX_cpd10516_e': 1000}
With the medium set, we can now simulate growth in these conditions:
In [7]:
from medusa.flux_analysis import flux_balance
fluxes = flux_balance.optimize_ensemble(ensemble,return_flux='bio1')
In [16]:
# get fluxes for the first 10 members
fluxes.head(10)
Out[16]:
| bio1 | |
|---|---|
| Staphylococcus aureus_gapfilled_18 | 14.890551 |
| Staphylococcus aureus_gapfilled_477 | 12.218825 |
| Staphylococcus aureus_gapfilled_430 | 19.198765 |
| Staphylococcus aureus_gapfilled_735 | 14.875922 |
| Staphylococcus aureus_gapfilled_916 | 12.223456 |
| Staphylococcus aureus_gapfilled_983 | 19.375070 |
| Staphylococcus aureus_gapfilled_371 | 13.113148 |
| Staphylococcus aureus_gapfilled_255 | 12.223456 |
| Staphylococcus aureus_gapfilled_729 | 14.891239 |
| Staphylococcus aureus_gapfilled_925 | 19.198765 |
In [10]:
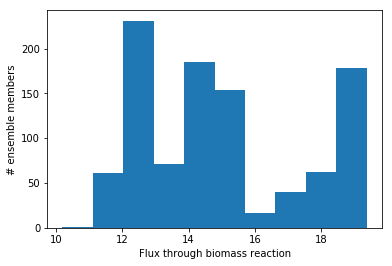
import matplotlib.pylab as plt
fig, ax = plt.subplots()
plt.hist(fluxes['bio1'])
ax.set_ylabel('# ensemble members')
ax.set_xlabel('Flux through biomass reaction')
plt.show()
You may want to perform simulations with only a subset of ensemble
members. There are two options for this; either identifying the desired
members for simulation with the specific_models parameter, or
passing a number of random members to perform simulations with the
num_models parameter.
In [14]:
# perform FBA with a random set of 10 members:
subflux = flux_balance.optimize_ensemble(ensemble, num_models = 10, return_flux = "bio1")
subflux
Out[14]:
| bio1 | |
|---|---|
| Staphylococcus aureus_gapfilled_300 | 18.441010 |
| Staphylococcus aureus_gapfilled_181 | 14.875922 |
| Staphylococcus aureus_gapfilled_667 | 17.618230 |
| Staphylococcus aureus_gapfilled_668 | 14.875922 |
| Staphylococcus aureus_gapfilled_639 | 14.186860 |
| Staphylococcus aureus_gapfilled_636 | 14.186860 |
| Staphylococcus aureus_gapfilled_738 | 14.643953 |
| Staphylococcus aureus_gapfilled_68 | 12.223456 |
| Staphylococcus aureus_gapfilled_87 | 14.875922 |
| Staphylococcus aureus_gapfilled_580 | 12.223456 |
In [15]:
submembers = [member.id for member in ensemble.members[0:10]]
print(submembers)
subflux = flux_balance.optimize_ensemble(ensemble, specific_models = submembers, return_flux = "bio1")
subflux
['Staphylococcus aureus_gapfilled_892', 'Staphylococcus aureus_gapfilled_851', 'Staphylococcus aureus_gapfilled_501', 'Staphylococcus aureus_gapfilled_927', 'Staphylococcus aureus_gapfilled_875', 'Staphylococcus aureus_gapfilled_500', 'Staphylococcus aureus_gapfilled_751', 'Staphylococcus aureus_gapfilled_849', 'Staphylococcus aureus_gapfilled_372', 'Staphylococcus aureus_gapfilled_421']
Out[15]:
| bio1 | |
|---|---|
| Staphylococcus aureus_gapfilled_372 | 12.223456 |
| Staphylococcus aureus_gapfilled_421 | 13.113148 |
| Staphylococcus aureus_gapfilled_500 | 19.198765 |
| Staphylococcus aureus_gapfilled_501 | 12.223456 |
| Staphylococcus aureus_gapfilled_751 | 14.209162 |
| Staphylococcus aureus_gapfilled_849 | 12.224814 |
| Staphylococcus aureus_gapfilled_851 | 19.375070 |
| Staphylococcus aureus_gapfilled_875 | 17.872504 |
| Staphylococcus aureus_gapfilled_892 | 19.375070 |
| Staphylococcus aureus_gapfilled_927 | 19.198765 |
Flux Variability Analysis¶
In [ ]:
Gene and Reaction Deletions¶
In [ ]: